Research
We work on two main projects: to elucidate the mechanisms of nutrient acquisition by Escherichia coli in the intestine and to use single-nucleotide-resolved transcriptome data for comparative gene expression analysis of competing intestinal bacteria.
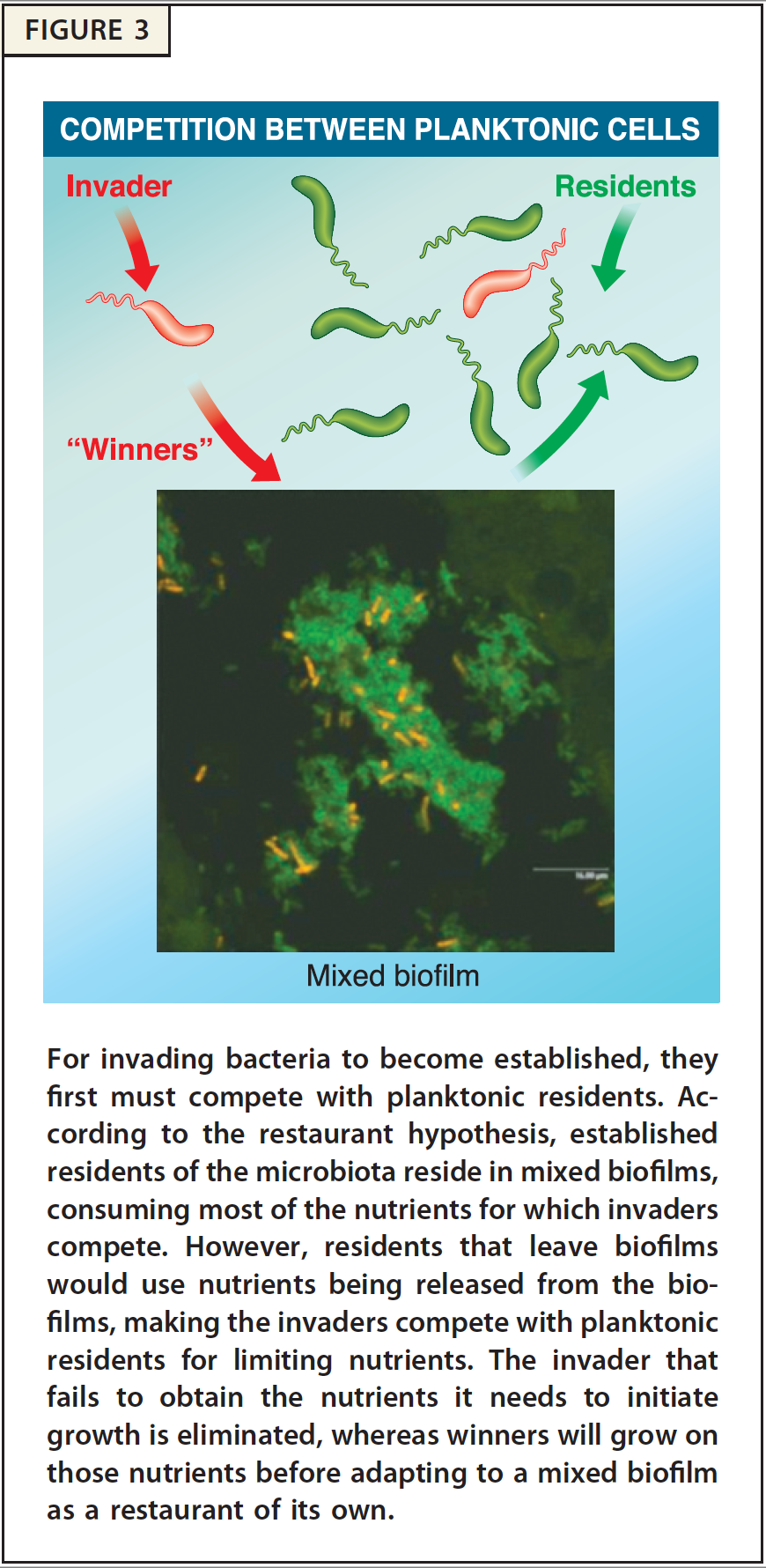
Mechanisms of Nutrient Competition in the Mammalian Intestine. This project addresses how E. coli colonizes the large intestine. The data illuminate cellular processes -- nutrition, stress-tolerance, and virulence factors -- that are important for colonization and pathogenesis. Importantly, we found that different E. coli biotypes compete for different sugars in the intestine, suggesting a mechanism for the succession of E. coli strains in healthy individuals and infection by E. coli pathogens that must overcome the colonization resistance barrier imparted by the resident E. coli. In addition, we learned that E. coli must respire oxygen to be competitive in the intestine. This finding suggests that E. coli scavenges oxygen to create anaerobic conditions that favor growth of the anaerobes that dominate the intestinal microbiota. We explore and characterize the symbiotic relationship between E. coli and the anaerobes that degrade complex polysaccharides, which in turn release simple sugars to cross-feed E. coli. Our results are explained by the restaurant hypothesis, which predicts microhabitats within the colonic mucus layer where facultative anaerobes, like E. coli, grow on sugars that are served locally by polysaccharide degrading anaerobes.
E. coli Gene Expression Database
Single-nucleotide Resolution Transcriptome Analysis. The goal of this project is to understand how bacterial cells work, from genome to transcriptome to metabolome, and to know their genetic circuitry and metabolic networks. To that end, work in my laboratory centers on how colonized bacterial cells grow and compete for nutrients in the microbial community of the mammalian intestine. Two major projects in the laboratory, both focused on E. coli, involve elucidating its symbiotic relation with the anaerobic microbiota in a mouse model of intestinal colonization and generating a single-nucleotide-resolution transcriptome map. Our RNA-Seq transcriptome map builds on the first published E. coli DNA microarray paper and the world’s largest E. coli gene expression database (GenExpDB). In the bioinformatics arena we developed a database schema and middleware, interfaced to intuitive displays, as a computational environment that we have used for discovery of genetic circuits controlling the general stress response and stringent response.
GenExpDB is hosted by Oklahoma State University!
GenExpDB is the world’s largest repository for E. coli gene expression data. This site is a widely used public resource for gene expression analysis.
Research Funding
1P20GM152333-01 — 2/1/2024 to 1/31/2029
NIGMS Institutional Development Award, Phase I COBRE — $10,742,999
Principal Investigator and Director
“Oklahoma Center for Microbiome Research”
RO1 GM117324 -01A1 — 3/1/17 to 1/31/24
NIH National Institute of General Medical Sciences — $1,181,670
“Mechanisms of Nutrient Competition in the Intestine”
The major goal of this project is to determine mechanisms of nutrient competition
between E. coli strains in a mouse model of intestinal colonization.
Diversity Supplement to parent project “Mechanisms of Nutrient Competition in the
Intestine”
Previous Research Funding
1R01GM095370-01 — 01/01/11 to 12/31/14
NIH NIGMS — $1,650,000 direct cost
“Symbiosis of E. coli and the Intestinal Microbiota in a Mouse Model”
RC1GM09207 — 09/30/09 to 8/31/12
NIH NIGMS — $1,000,000 direct cost
“Comprehensive Mapping and Annotation of the E. coli Transcriptome”
2RO1-AI48945-05 — 6/01/04 to 5/31/09
National Institutes of Health — $2,158,563 total cost
“Growth and Colonization of the Intestine by E. coli”
More than 25 grant awards from DOE, USDA, NREL, NSF, Genencor and NIH continuously funded since 1990, totaling more than $28 million.